Service
LiLac-QSPLiLac-QSP 플랫폼 이식 서비스 모델
약물 노출 및 반응 예측을 통한 상관성 확보는 신약개발 전주기에 영향을 주는 요소로 회사 내부 의사 결정 및 전세계 의약품 허가관련 기관(예, 미국 FDA, 유럽 EMA, 한국 식약처 등)에서 임상시험 및 허가 관련 결정 시 지원하는 핵심 자료로 분류되어 제출되고 있음.
따라서, 신규 타겟 메커니즘의 이해에 대한 지원, 선도물질 및 신약후보물질의 약물 노출-반응 비교 평가를 통한 우선순위 지원, 초기 및 후기 임상시험 디자인 최적화 지원, 약물 노출에 대한 흡수, 분포, 대사, 배설 특성 파악을 통한 환자군간 변동성 확인 지원, 약물 반응에 대한 효용 대비 위험비 분석을 통한 약물 사용 유용성 평가 지원, 기존 약물 요법 과의 비교 평가를 통한 신규 약물요법 반응에 대한 비교 평가 지원, 임상시험 수행 불가능한 특수 환자군(소아, 신장애 환자 등)에 대한 약물 노출-반응 예측 지원
의뢰사
- 제약회사로 부터 약물
- 노출-반응 상관성 확보 의뢰

LiLac
- LiLac-QSP 플랫폼 활용
- 노출-반응 상관성 확보 결과 도출 및 인계

의뢰사-LiLac
- 인계된 결과에 대해 LiLac_QSP 플랫폼을 활용하여 확인 및 결과 환류

의뢰사-허가기관
- 최종 확정된 결과를 기반으로 하여 허가 관청에 제출
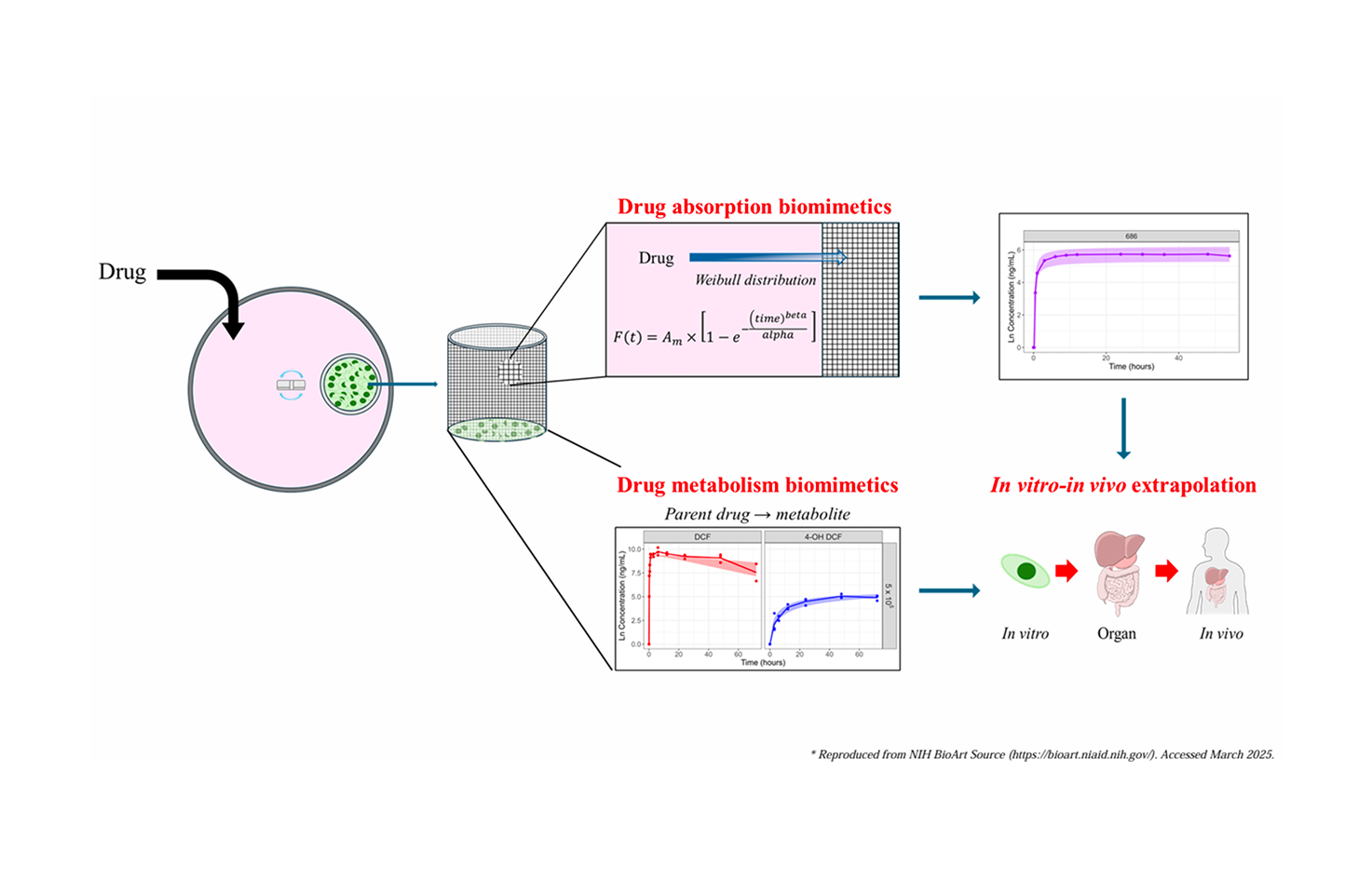
PK simulation platform for biomimetic in vitro system
This platform is an interactive R Shiny-based web application designed to predict in vivo drug kinetics by integrating a patented biomimetic in vitro system (Korean Patent No. 10-2022-0153547) with mechanistic pharmacokinetic modeling. Leveraging experimental data on drug diffusion and cellular metabolism from the porous mesh-based system , the platform simulates the complex processes of drug absorption and metabolic clearance. This model-based strategy aims to provide a reliable, animal-free tool for pharmacokinetic prediction, thereby supporting the 3R (replacement, reduction, and refinement) principle by reducing the reliance on animal testing in drug development.
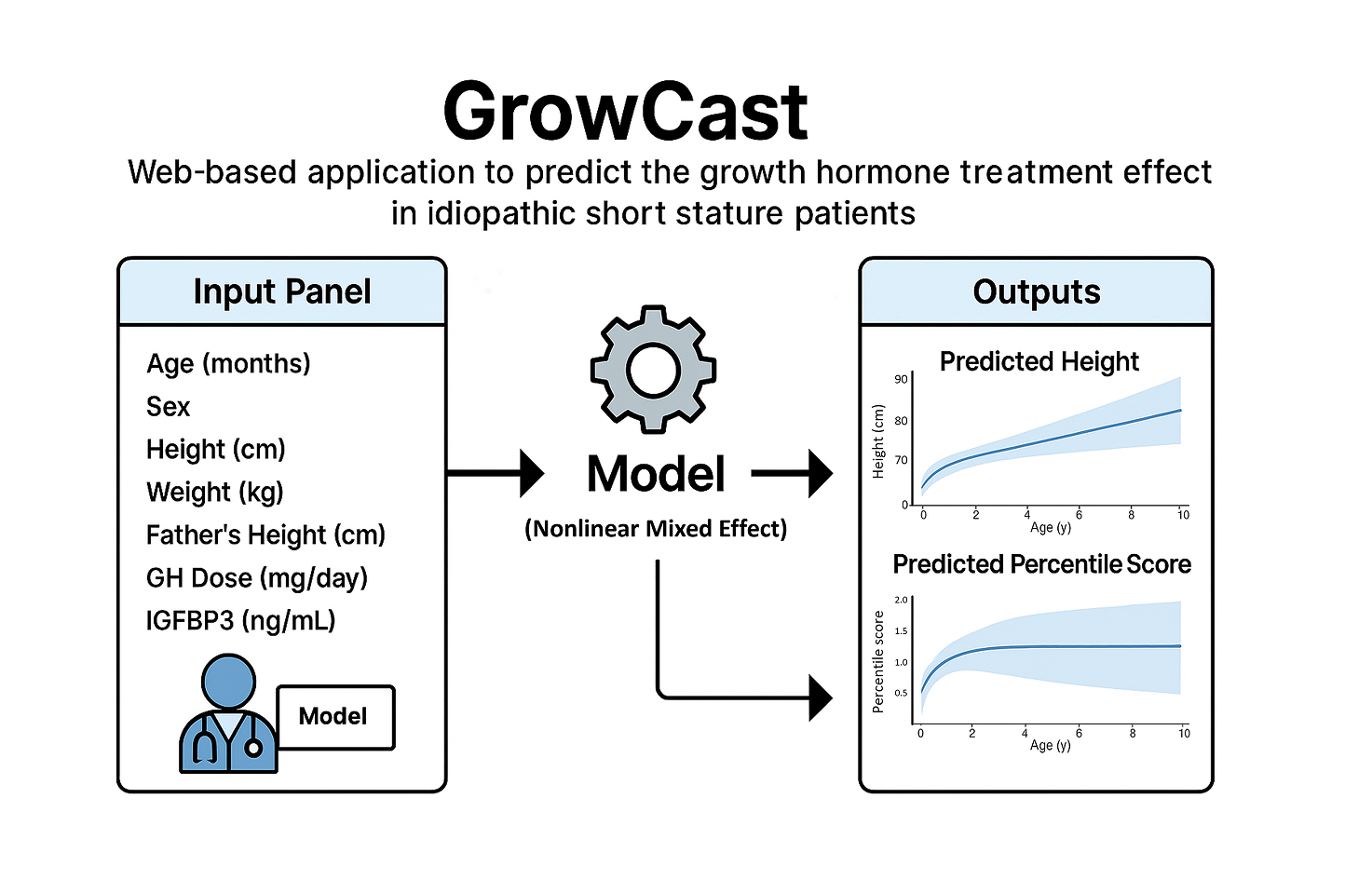
GrowCast
GH Effect Predictor for Idiopathic Short Stature Patients
GrowCast is a web-based R-Shiny application that predicts the growth hormone (GH) treatment effect in patients with idiopathic short stature (ISS). By entering patient information and treatment parameters, the app provides visual forecasts of expected height trajectories and percentile changes. The model applies a Nonlinear Mixed Effect model to deliver interpretable and accurate predictions. The output displays predicted height curves (with uncertainty intervals) and percentile scores in interactive graphs. With its intuitive UI and instantly reactive interface, users can easily assess the expected treatment outcomes.
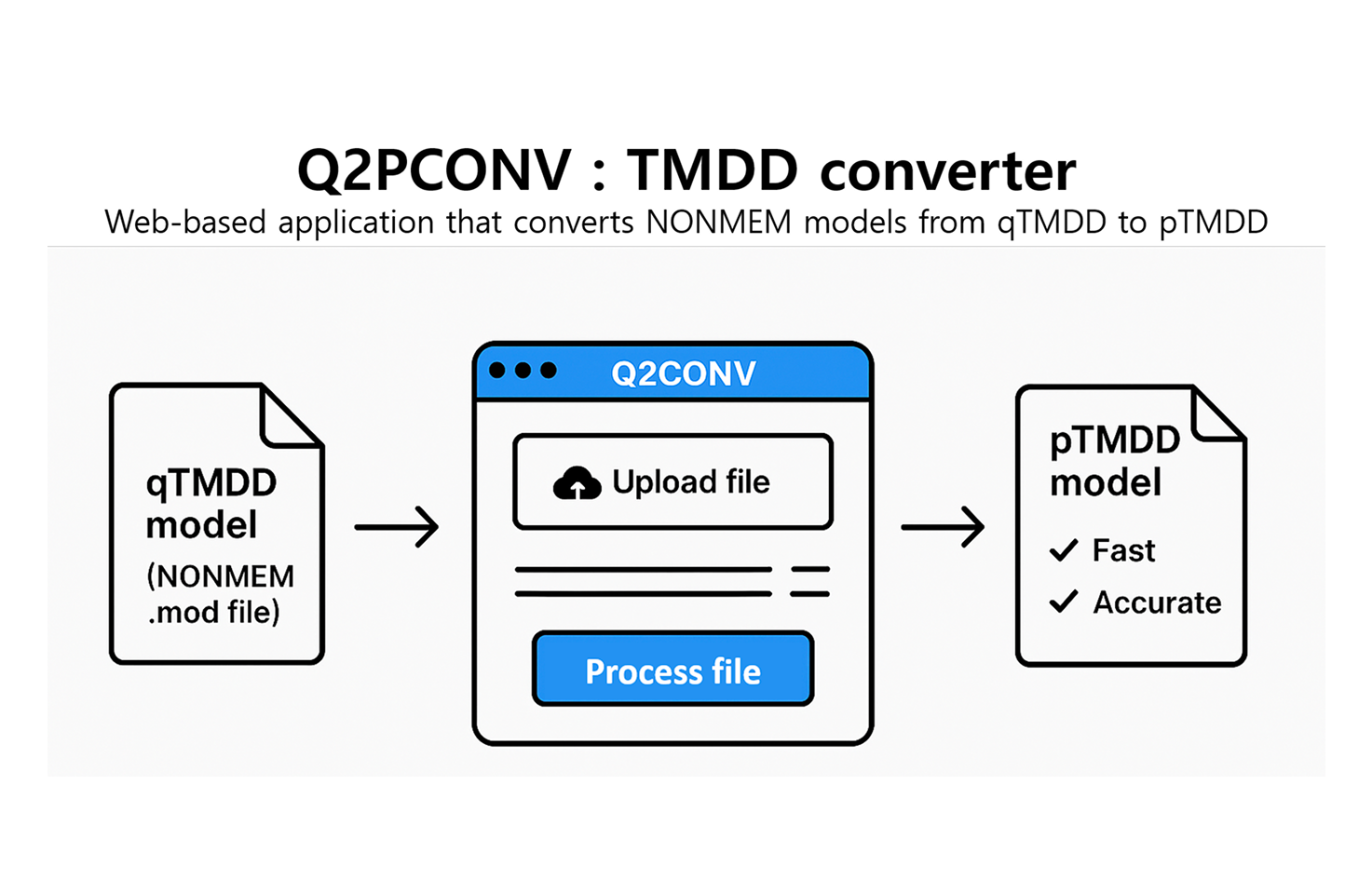
Q2PCONV (Convert TMDD)
Q2PCONV is a web-based R Shiny tool that converts qTMDD models into the newly developed pTMDD approximation. The pTMDD model, derived from the qTMDD model via first-order Taylor expansion, provides higher accuracy than traditional Michaelis-Menten models while being faster to compute than the Quasi-Steady-State (qTMDD) approach. Users can upload NONMEM .mod files and specify options such as receptor type, observation type, and receptor compartment. With a single click, the tool processes and translates the qTMDD model into pTMDD format. The converted file can then be reviewed and downloaded directly from the application.